Research
Our research question: What keeps the Neurospora genome transposon-free?
Our research interest:
The fungus Neurospora crassa possesses an exceptionally effective ability to defend its genome against the proliferation of transposons and other repeated DNA. Only one Neurospora strain, of >1000 examined, was found containing transposons. This strain, isolated from Adiopodoume in West Africa, hosts the retrotransposon Tad (Transposon of Adiopodoume). All the other Neurospora strains examined contained only relics of Tad that were inactivated by numerous G:C to A:T mutations. No other transposon was found in Neurospora. The transposon-free genome of Neurospora attests to the effectiveness of its genome defence processes. Our research focuses on two of these processes, RIP (repeat-induced point mutation), and MSUD (meiotic silencing by unpaired DNA), also known simply as meiotic silencing.
RIP targets G:C to A:T hypermutation to repeated DNA sequences during a sexual cross. We developed an assay for RIP, and used it to show that the RIP machinery is titrable by chromosome segment duplications (Dp). These studies also led us to define the breakpoints of several Dp-generating chromosome rearrangements onto the genome sequence. Using the RIP assay we showed that the Adiopodoume strain can exert a dominant RIP suppressor phenotype. The Adiopodoume strain contains ~40 copies of Tad, each ~7 kb, therefore this ~280 kbp of duplicated DNA might be contributing to titration of the RIP machinery, whereas in low copy strains Tad remains vulnerable to RIP.
Meiotic silencing by unpaired DNA (MSUD) is an RNAi-mediated elimination of the transcripts of any gene that is not properly paired with a homolog in meiosis. MSUD can be demonstrated in crosses of the standard laboratory OR strains with the ::Bmlr and ::mei-3 tester strains. Silencing of the bml (β-tubulin) and mei-3 genes in these crosses results in dramatic ascus-development abnormalities. MSUD does not occur in homozygous tester A x tester a crosses, nor in crosses of the testers with the semi-dominant suppressors of MSUD (eg., Sad-1 and Sad-2), and the asci develop normally. The suppressor alleles are presumed to prevent the proper pairing of their wild-type homologues, and thus induces them to autogenously silence themselves.
Wild-isolated N. crassa strains could be classified into three types based on the phenotype of their crosses with the testers. In crosses with “OR” and “Sad” type strains the Bmlr and mei-3 genes are, or are not, silenced. Whereas in crosses with “Esm” type bml was silenced but not mei-3+. Presumably, bml is more sensitive to silencing than mei-3, and sequence polymorphisms between the genomes of the OR-derived testers and the Sad and Esm strains might cause one or more gene essential for meiotic silencing to become unpaired and silence itself, thus shortening the duration of silencing. Thus, MSUD could become very fleeting in the cross with Sad type, of intermediate duration in the cross with Esm type, and persist throughout the cross only with OR type. We are now trying to determine whether MSUD will occur if a new tester made in the genetic background of a Sad type strain is used to perform a cross that is heterozygous for the tester allele but isogenic for the rest of the genome. In the course of developing the new Sad-derived strains we found that the Sad versus Esm difference can have a genotype-independent basis.
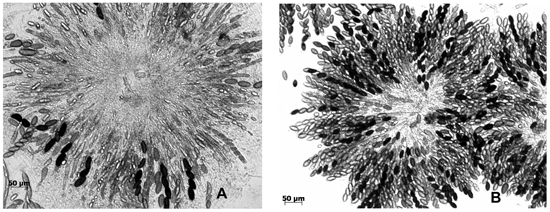
Figure 1. Varied phenotypes in crosses of the N. crassa ascus-dominant Dip-1 mutant with two wild-isolated strains. Left, a rosette of asci from the cross Dip-1 a x Roanoke-1m A shows the mutant phenotype, namely, several asci with two to four large ascospores instead of the normal eight. Right, almost all asci are eight-spored in the rosette from the cross Dip-1 a x Klong Rangsit A.
|