|
|
|
| Home » Neuroscience and Cell Biology » Research |
| Neuroscience and Cell Biology |
Research
For more information click here to visit PI's personal website.
[Please note that the link is outside of CDFD domain]
The long term goal of my group is to understand the development of central nervous system (CNS) using model organism Drosophila melanogaster (fruit fly).
The key question is how various cellular phenomenon’s like cell quiescence, proliferation, differentiation and apoptosis are coordinated for neural stem cells (NSC) during development. This is important to generate a variety of different cell types and cell numbers which are required for the formation of a functional CNS.
Coordination of these phenomenon is a consequence of an interplay between intercellular signalling (amongst different cell types of CNS), as well as cell autonomous epigenetic regulation of genes required for cell proliferation, differentiation and apoptosis. We employ a multi-disciplinary approach including cell biology, genetics and biochemistry to address these questions in flies.
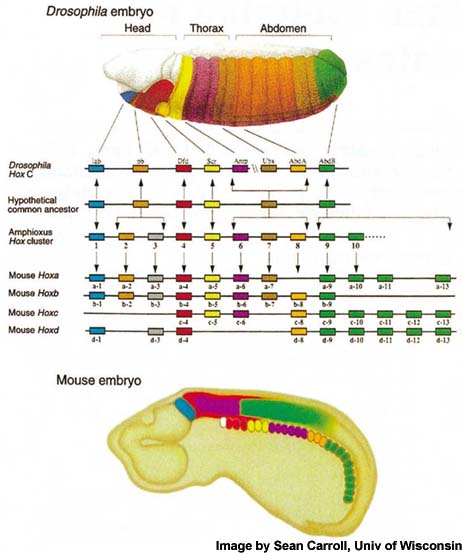
One of the earliest steps in CNS development of multicellular organisms is the specification of anterior posterior (AP or head to tail) axis, which is regulated by Hox family of transcription factors. Hox genes are highly conserved homedomain (HD) containing factors (Fig-1) which are expressed in CNS (Fig-2) of both insects as well as vertebrates.
|
 |
|
Figure-1.Drosophila and vertebrate Hox genes show conserved expression along the AP axis of body. Paralogs of a Hox protein in two different species are shown by same color. There are 8 Hox genes in Drosophila and 39 Hox genes in vertebrates (divided into 4 clusters) as shown (Image by Sean Carroll, Univ of Wiscosin, Madison).
|
|
|
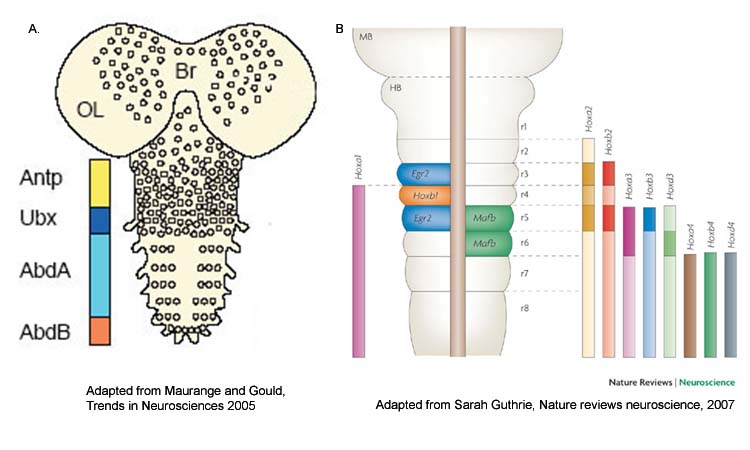 |
| Figure-2. Hox genes are expressed in CNS of Drosophila (A) and vertebrates (B) where they play a role in AP axis determination Adapted from Maurange and Gould, Trends in Neurosciences 2005 and Sarah Guthrie, Nature reviews neuroscience, 2007. |
|
Hox genes are suggested to give NSCs their positional identity, which eventually determines how many times NSCs in a specific region of CNS will divide; when they will exit cell cycle, or undergo apoptosis; and what will be the different kinds of cell types their progeny will differentiate into (neuronal or glial cell types).
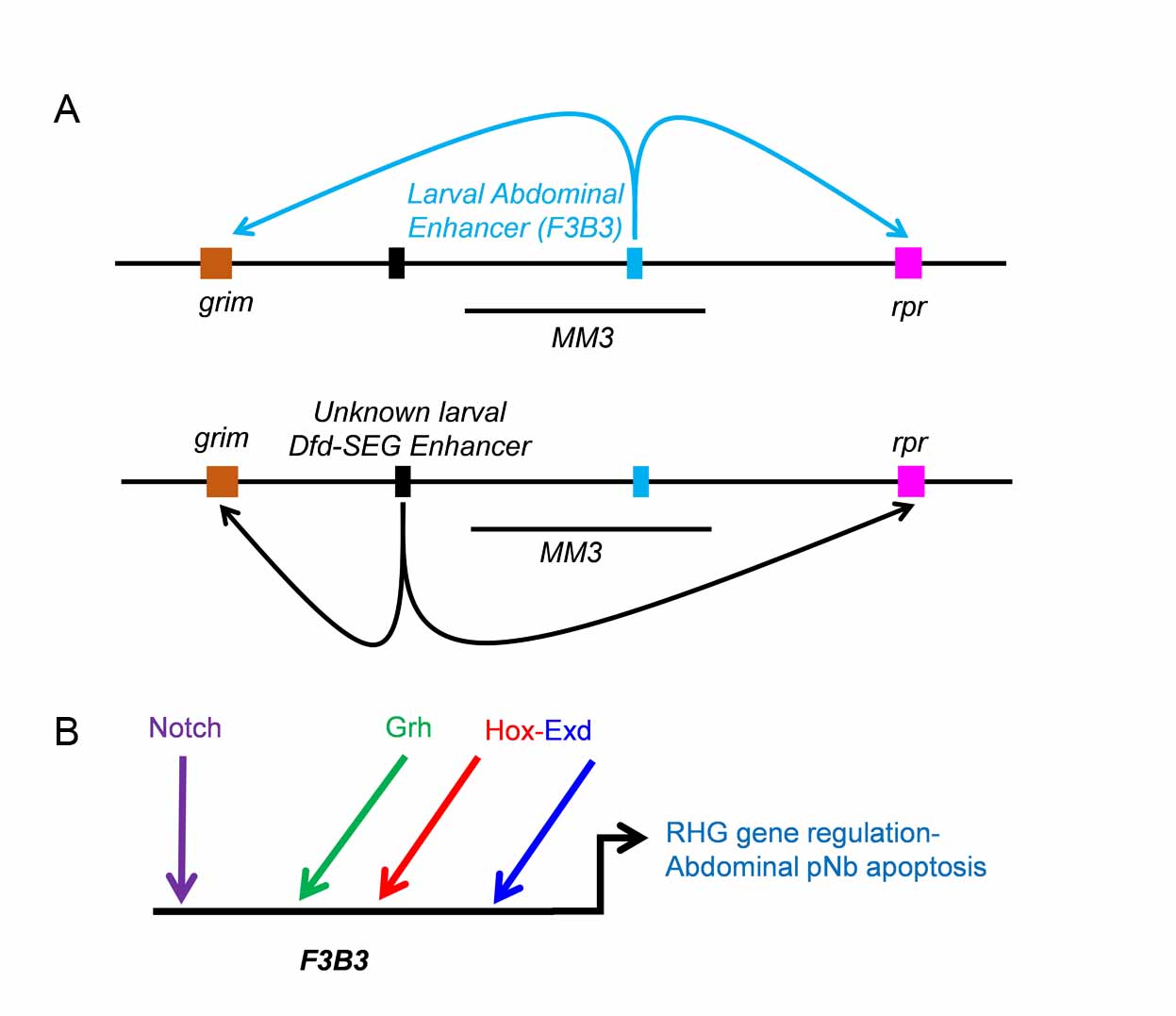
Our most recent work has shown that apoptosis of NSCs in different regions of developing CNS are carried out by Hox genes. Hox gene executes this apoptosis along with a combination of common transcription factors and Notch signalling pathway. Interestingly, we find that these factor employ different region specific enhancers to activate apoptotic genes and cause NSC’s cell death (Khandelwal et al., PloS Genetics, 2017, Fig-3).
The choice of transcription factors and signalling pathway is determined by developmental history of the stem cell lineage, more specifically its location in developing CNS.
Understanding some of these problem will give insights into a broader question of how cell proliferation and fate determination is coordinated with overall development of the animal in general.
Since the underlying principles of CNS patterning are conserved across species and both mammalian and Drosophila NSCs have very similar molecular properties (like progressive lineage restriction, mitotic quiescence and asymmetric cell division), therefore the studies done in Drosophila will be relevant in a wider context
|
 |
|
Figure-3. TFs and Notch signaling in NSC apoptosis. (A) Regulation of apoptotic genes grim and reaper in abdominal and Dfd-SEG region of CNS happen through two distinct enhancers. Enhancer for Dfd-SEG is yet to be identified but lies outside 55Kb genomic deletion MM3 and is arbitrarily shown 5’ to MM3. (B) Model for regulation of RHG genes in abdominal NSC. bHLH TF Grainyhead, Hox, Extradenticle (Exd) and Notch signaling play a direct role in regulation of RHG genes through 1Kb F3B3 enhancer. The site of origin of the activating ligand for Notch signal is yet to be established conclusively (Khandelwal et al., 2017)
Suggested Reading
For introductory developmental biology and AP axis specification in flies and vertebrates
Any standard developmental biology textbook is a good point to start, below I list a few:
The Chapter on developmental biology in “Molecular Biology of cell-by Bruce Alberts”
The chapter on Drosophila axis formation from “Principles of Development-by Lewis Wolpert” and “Developmental Biology-by Schott Gilbert”
The Making of a Fly: The Genetics of Animal Design, by Peter A. Lawrence
To learn about Drosophila development, go to http://flymove.uni-muenster.de/ click on processes tab, the website gives a concise and easy overview of fly development.
To know more about axis determination during central nervous system development
Reichert, H. A tripartite organization of the urbilaterian brain: developmental genetic evidence from Drosophila. Brain Res Bull 66, 491-494, (2005).
Maurange, C. & Gould, A. P. Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci 28, 30-36 (2005).
Alexander, T., Nolte, C. & Krumlauf, R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol 25, 431-456, (2009).
Working in this lab is subject to your clearing appropriate CDFD selection procedures (for details contact EMPC Section, CDFD: academia<at>cdfd.org.in). At the moment there are no postdoctoral positions available but anyone interested is encouraged to get in touch.
A write up of 250 words explaining your specific academic interest in the lab is expected from everyone seeking to explore the possibility of working in this lab at any position. A prior knowledge in genetics and biochemistry will be advantageous but not a pre-requisite.
|
|
| Last updated on : Friday, 3rd June, 2022 |
|
|
|
|